DNA expression and regulation– protein synthesis
DNA, RNA and ribosomes, in that order, are essential components in the synthesis of proteins. DNA contains the information necessary not only for reproduction, but also for daily cell growth and maintenance. Messenger RNA carries the information to the ribosomes. With the help of yet another kind of RNA, the ribosomes assemble the proteins. All this depends on gene regulation.
The use of DNA to initiate protein synthesis is called DNA expression.
This sequence of events is summed up in the so-called central dogma of molecular biology, often paraphrased as “DNA makes RNA and RNA makes protein.” More precisely, DNA is transcribed inside the nucleus to make mRNA, which is expelled from the nucleus to the cytoplasm, where it is translated to protein by ribosomes.
DNA –> transcription (nucleus)→ mRNA→ translation (ribosome)→ protein.
The recipe is expressed in “bytes” of three nucleobases; one three-base byte is referred to as a code-word. When transcribed to its complementary form in mRNA, it is called a codon. Since each base can have one of four values (C, G, A or T, in DNA), the codon can take on 64 values.
RNA transcription
The enzyme which does the work of “reading” a gene on the DNA and building a corresponding gene of RNA is called RNA polymerase[ref]In fact, there are several forms of RNA polymerase, but that complexity is well beyond the scope of this document.[/ref]. There are at least four types of RNA and transcription makes them all. For protein synthesis, the RNA constructed is called mRNA, or messenger RNA. The DNA recipe begins with a sequence called the promoter. RNA polymerase contains a complementary sequence which binds to the promoter and launches transcription. As will soon be seen, transcription is started only if it is allowed by gene regulation. RNA polymerase unwinds a part of the DNA chain and reads code-words, starting with the promoter. As it reads the DNA, it constructs a complementary chain, called pre-mRNA, from nucleotides. It is complementary in the sense that if the DNA contains a C (or A or G or T) then the pre-mRNA contains a G (or U or C or A – remembering that RNA replaces T by U).
The raw materials RNA polymerase uses to construct RNA are nucleoside triphosphates (NTPs). An NTP molecule has two of its phosphate groups which contain a significant amount of energy from ATP. This energy is used to bond the nucleotides together to form RNA.
The RNA polymerase moves along the DNA, unwinding sections as it goes, reads the code words and assembles the appropriate pre-mRNA codons from NTPs. The separated DNA strands recombine in its wake. Eventually, it reaches a transcription-terminator sequence in the DNA and ends transcription. It now has gone through three steps, known as initiation, elongation (of the produced pre-mRNA) and termination. The pre-mRNA then is released into the nucleoplasm.
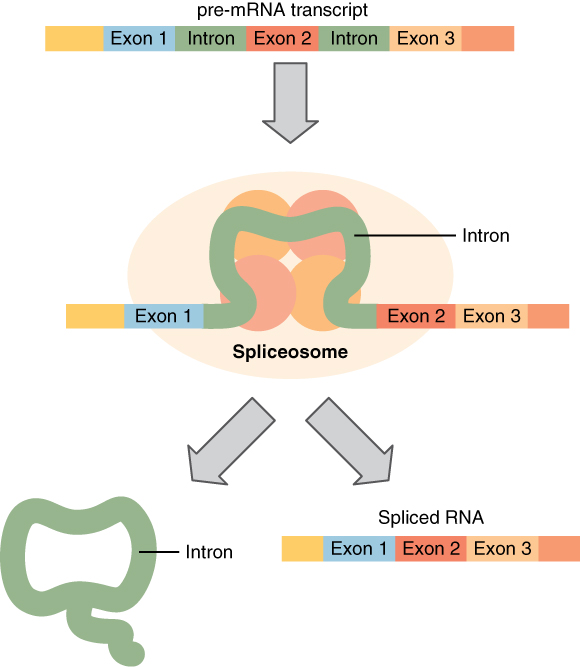
Splicing mRNA, from Openstax College
Before leaving the nucleus, the pre-mRNA must be cleaned up. This is needed because DNA contains non-coding, or junk, sequences. The codons which should be kept are called exons (like “expressed”) and those which should be deleted are called introns (like “interrupted”).[ref]I would have preferred for exon to mean “exclude” and intron to mean “include”, but some contrary biologist decided otherwise. He could at least have taken a vote![/ref] Small particles called “snurps” (for snRNPs, or small nuclear ribonucleoproteins), made up of RNA and proteins, bind together to form spliceosomes, which remove introns and splice the exons back together again, resulting in a cleaned-up form of mRNA.[ref]Are you wondering how the snurps can recognize the introns an exons? So am I. All I can say is that it is quite complicated and has something to do with methylation of the DNA strands. It is currently not completely understood why there are introns at all, but there are indications that they may be of importance.[/ref]
The mRNA is then moved out of the nucleus for the next step.
Protein synthesis – translation
After the mRNA leaves the nucleus, it is used to provide the input data for the synthesis of proteins. This takes place on ribosomes.
There are two sorts of ribosomes in eukaryotic cells, depending on their location.
- Free ribosomes float in the cytoplasm and make proteins which will function there.
- Membrane-bound ribosomes are attached to the rough endoplasmic reticulum; they are what makes it look “rough”. Proteins produced there will either form parts of membranes or be released from the cell.
In most cells, most proteins are released into the cytoplasm.
Ribosomes are made of ribosomal RNA, or rRNA (one more kind of RNA), and proteins. They are constructed within the nucleolus as two subunits, which are released through the nuclear pores into the cytoplasm.
In addition to the mRNA and the ribosome subunits, a method Is needed for supplying the appropriate amino acids to be linked by peptide bonds to make up the protein or enzyme being constructed. Enter still one more kind of RNA, transfer RNA, or tRNA.
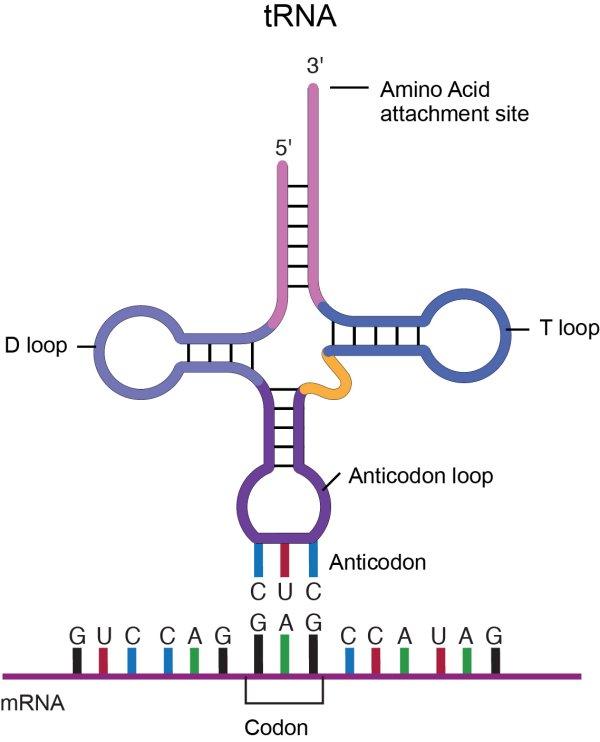
Transfer RNA, tRNA, from National Human Genome Research Institute
A molecule of tRNA is a molecule of RNA folded into a double strand with loops which give it a precise 3-dimensional shape. The loop on one end has an anticodon, the function of which is to match its complement codon on mRNA. The other end has a binding site (adenylic acid) for a specific amino acid. So the tRNA is the “dictionary” which converts codons into amino acids[ref]This of course poses the question, where do the tRNA molecules come from? Good question.[/ref]. A tRNA molecule is “charged” with an amino acid molecule by a tRNA-activating enzyme which uses energy from ATP to covalently bond the appropriate amino acid from molecules in the cytoplasm. Such tRNA molecules, carrying an amino acid, are called aminoacyl tRNA.
The ribosome itself contains three assembly areas or spaces called, in order of occupation, the A-site, the P-site and the E-site. Initially, the ribosome subunits are floating independently in the cytoplasm or attached to the RER. The initiation of translation begins when the small subunit binds at its P-site to the START codon of the mRNA strand. Then the corresponding tRNA (methionine) binds to the START codon and the large ribosome subunit is attached, completing assembly of the ribosome. Now the first tRNA is in the P-site and next mRNA codon in the A-site. The methinone constitutes the beginning of the peptide chain which will become the protein. Then a cycle takes place in which the ribosome reads in the mRNA strand, like computers of my youth read in paper tape, each new codon arriving in A.
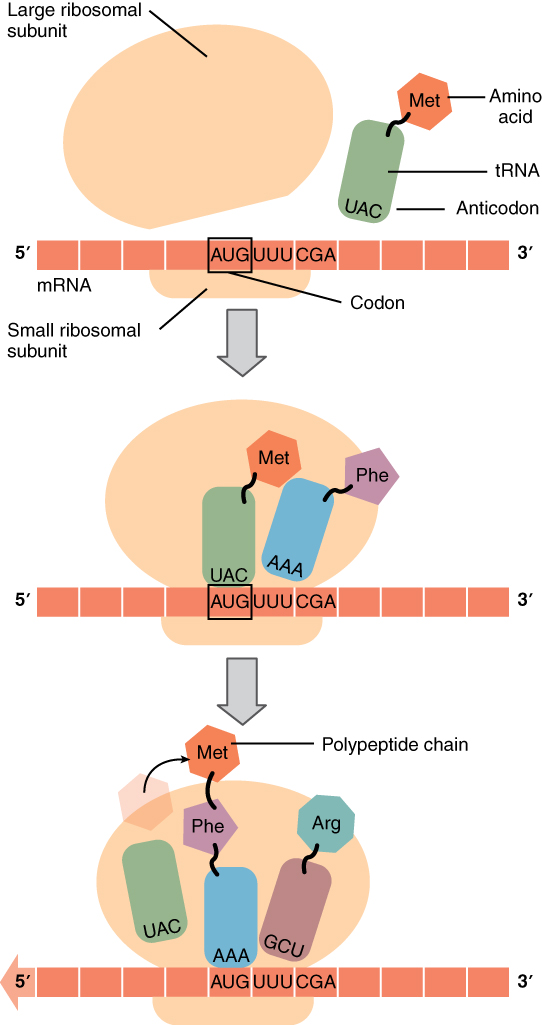
Gene translation in the ribosome, from Openstax College
The process then pursues the elongation stage of translation. The aminoacyl tRNA for the codon in the A-site is carried in, so the first two amino acids are now in the P and A sites. The ribosome then catalyzes the formation of a peptide bond between these two amino acids. The ribosome then moves the mRNA so the P-site amino acid enters the E-site, the A-site one enters the P-site, and a new one enters the A-site. It continues like that until a STOP codon enters the A-site and brings about termination of translation and release of the completed peptide chain.
All these steps of transcription and translation require energy, so protein synthesis is one of the most energetically costly of cell processes. Much of this energy is used to make enzymes essential to the functioning of the cell. Most enzymes are proteins.
Once part of a strand of mRNA has left one ribosome, it can enter another. One strand may actually be in 3 to 10 ribosomes at once, in a different step of translation in each one. Such clusters of ribosomes translating the same mRNA strand are called polyribosomes.
Regulation of gene expression
Every cell in an organism has the same complete genome in its nucleus and so has access to all the same protein “recipes”. But, for example, heart cells should not produce proteins used only by the liver and no cell should produce proteins in quantities beyond what it can use. Cells change over time too: Think of the adaptation to pregnancy or disease.
Cells are specialized and so express different genes: Differential gene expression leads to cell differentiation. Controlling which proteins to express and when is called regulation. Note that this is one more instance of communication in the body, telling genetic machinery what to express when.
Regulation of prokaryotic cells
Regulation in prokaryotic cells is relatively simple, as there is no nucleus. so transcription and translation take place almost in the same place and at the same time. Regulation in prokaryotic cells, though, almost always concerns transcription.
An example from a prokaryotic cell will show how this works – and introduce some new terminology.
The bacterium E. Coli normally uses glucose for energy. But if glucose is absent and lactose is present, it can use the lactose. Bacteria arrange groups of genes to be controlled together into a structure called an operon. The set of proteins necessary for the use of lactose are part of the lac operon. The operon begins with a promoter, which indicates the beginning of the operon and is the site where RNA polymerase binds to begin the transcription. In between the promoter and the set of genes, of which there may be any number, is a sequence called the operator, which is where DNA-binding genes bind to regulate transcription[ref]Look out for the terminology: Sean B. Carroll refers to the operator as a genetic switch, a term we will meet with in the disussion of regulation in eukaryotes.[/ref]
When no lactose is present, a protein called the lac repressor is bound to the operator and the state of the lac operon is “off”. (Figure.) This is because the repressor blocks access to the rest of the operon.
The gene for the lac repressor is a constitutive gene: It is always expressed because it is the recipe for an essential protein. On the other hand, a regulated gene, is expressed selectively.
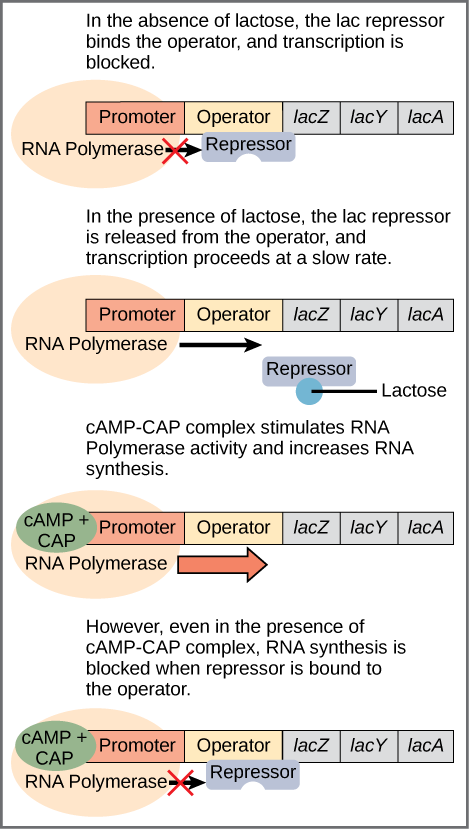
Regulation of the lac operon, from Openstax College
In addition to the binding site for the operator, the lac repressor has a second, allosteric, binding site. When lactose is present, an isomer of lactose binds to the allosteric site of the repressor, which causes it to change its form and unbind from the operator. The lac operon is now in the “on” state. This form of regulation is called induction: Lactose is said to be the inducer of the lac operon and acts through the allosteric site of the lac repressor. Transcription now occurs, but slowly. Because some glucose still may be present, it is not certain that the proteins mapped by the lactose-digesting genes are needed. This depends on how much glucose is lacking.
The second part of this process depends on the presence of glucose and is regulated by a second DNA-binding protein, CAP (catabolite activator protein). CAP is also an allosteric protein with one DNA-binding site and one allosteric site which binds to cyclic AMP (cAMP). CAP is only active when it is bound to cAMP. You guessed it, cAMP levels are high when glucose levels are low.[ref]If we go one step back, we see that glucose binds to an allosteric site on the enzyme adenylate cyclase, which makes cAMP from ATP, and disables it. So lack of glucose stimulates production of cAMP, which binds to CAP, which binds to the promoter to enhance synthesis.[/ref] In that case, cAMP-CAP binds to the promoter and enhances transcription of the genes. So lactose can be considered the “on-off” switch for transcription of genes for lactose-digestion and cAMP-CAP, the “volume control”.
Regulation of eukaryotic cells
In eukaryotic cells, transcription occurs inside the nucleus and translation outside, so mRNA is shuttled across the nuclear membrane in between the two processes. Regulation in eukaryotic cells therefore may take place inside or outside the nucleus or at any step in the expression pathway, including control of access to the gene in the DNA, control of transcription, pre-mRNA processing, mRNA lifetime and translation, and modification of the final proteins. Even the activity levels of enzymes which facilitate expression can be controlled.
Pre-transcription regulation
Inside the nucleus, histones, around which chromatin is wound to make nucleosomes, can wind or unwind to change spacing of the nucleosomes and thereby allow or deny access to genes. This process is a form of epigenetic regulation.[ref]Look out, epigenetic regulation is used for somewhat different notions, too, and they are not all necessarily so.[/ref] Since histones are positively charged and DNA, negatively, modifying the charge by adding chemical “tags” to either modifies the configuration of the DNA.
Transcription regulation
The most frequent regulation of expression in eukaryotes is during transcription. Control of transcription by the prokaryotic lac operon is a relatively simple process: In order to begin transcription of a gene, RNA polymerase must bind with the gene’s promoter but cannot do so if a lac repressor is bound to the operator region which follows the promoter on the gene.
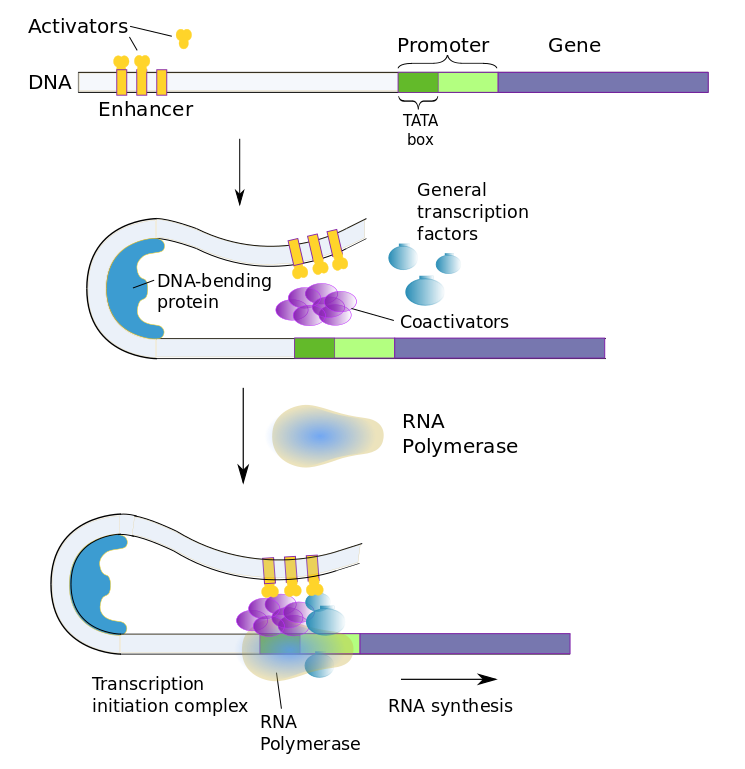
Eukaryotic gene transcription is regulated similarly, but with more of everything. Instead of a repressor which binds to an operon, eukaryotes have a slew of transcription factors which bind to multiple regulatory sequences. Regulatory sequences, sometimes referred to as switches, may be almost anywhere on the DNA strand, even far from the gene. The existence of multiple switches for each gene allows the gene to be present In different types of cells, but activated selectively in each by different switches.
There are two types of transcription factors.
- general transcription factors affect any gene in all cells and are part of the transcription-initiation complex;
- regulatory transcription factors affect genes specific to the type of cell.
The two types of transcription factors work together with three types of regulatory sequences (transcription-factor binding sites).
- promoter proximal elements are, of course, near the promoter and turn transcription on;
- enhancers are far from the regulated genes or in more than one place and also turn the transcription on;
- silencers are also far away from the regulated genes but turn transcription off.
Activator transcription factors are those which bind to enhancers to promote expression, repressor transcription factors, to silencers to decrease expression.
The promoter in eukaryotic cells is more complex too, The basal promoter begins with the TATA box, recognized by its beginning which contains the seven-nucleotide sequence TATAAAA, followed by a set of transcription-factor binding sites.
The whole set of transcription factors is summed combinatorially to determine whether or how much the gene will be expressed. Selective promotion or inhibition at combinations of these sites can therefore bring about tissue-specific gene expression. Each tissue type may have its own specific enhancer or silencer sequence for the same gene. For instance, the neuron-restrictive silencing element (NRSE) is a repressor which prevents genes from being expressed in any cells which are not neurons. In addition, environmental changes may bring about different gene expression according to current, perhaps temporary needs.
Coactivator proteins bind with general and regulatory transcription factors to form the transcription-initiation complex. RNA polymerase only binds to the transcription-factor complex.
The above figure shows the case of an enhancer bound by activator transcription factors.[ref]The bobby-pin curl is an idealization; DNA shapes are far more complex than that.[/ref] The enhancer, on the left, originally is quite far from the promoter, until DNA bending causes it to change its shape, allowing the enhancer to come in contact with the promoter and the rest of the transcription-initiation complex.Since transcription factors are proteins, they too are coded by genes and these genes are regulated in turn by other transcription factors. Eventually, it is the original cell (such as a fertilized egg) plus the environment which start the chain going. Of course, only genes which are present can be influenced; it’s nature and nurture.
Transcription factors and signaling elements coded by some of these genes make up the genetic toolkit, as we will see in a moment.
Splicing regulation
In between transcription and translation, proteins may interfere with spliceosomes to modify splicing of pre-mRNA. Different intron selections can allow different mRNAs to be produced from the same pre-mRNA, a phenomenon known as alternative splicing.
Pre-translation and translation regulation
RNA does not hang around forever, nor should it. Eventually, it is degraded and is no longer functional. So controlling its lifetime is another way of regulating its activity.
Yet another type of RNA, very short-stranded microRNA or miRNA, can bind with complementary mRNA before it is translated and signal that it should be destroyed by the cell. For this purpose, miRNA also associates with RISC (RNA-induced silencing complex).
Other proteins, RNA-binding proteins (RBPs), can bind with the 5′ cap or the 3′ tail of the mRNA and either increase or decrease its stability.
Phosphorylation or attachment of other chemicals to the mRNA protein initiator complex also inhibit translation.
Similar bindings may take place on the protein after translation and modify its stability, lifetime or function.
The development genetic toolkit – what evo devo tells us
Development, meaning embyronic development, is the process in which a genotype, a set of genes, becomes a phenotype, a particular living organism. Mutation works on genes, but natural selection works on phenotypes, the results of development. So evolution and development work hand in gene, so to speak, and the branch of biology which studies them together is called “evo-devo“.
The homeobox is a genetic sequence of DNA about 180 bases long. It is a sequence of DNA, of genetic material. It codes for about 60 amino-acid residues and these proteins are the homeobox domain, or the homeodomain. The reason the homeobox is really special is that it is “conserved” across most species of eukaryotes, meaning they all have similar homeobox sequences. Some such sequences are the same in frogs and mice by up to 59 of 60 base pairs. It is thought that there are about two dozen types of homeodomains, therefore of homeoboxes. The ubiquity of the sequence is fairly astounding in itself. It means that the sequence could not have evolved independently all those many times – think of the number of species concerned. So the homeobox must date back to be on the order of 500 millions years old.
But there’s more. The homeobox is not a gene by itself, but exists within many different, much larger genes – indeed, hundreds of times larger. Since they all contain the homeobox, they are called homeobox genes.
Many animals have a disposition of body parts along an axis, such as the antennae, wings and legs along the body axis of a fruit fly, or the existence or not of ribs along the vertebrae of a vertebrate animal. It turns out that the choice of body part at each segment along the axis is regulated by a transcription factor coded by a single gene – the Hox gene. Hox genes are an example of homeobox genes; they contain the homeobox sequence. These Hox “master” genes control the developmental differentiation of, for instance, a fruit fly’s serially homologous body parts;[ref]The front legs of a cat and our arms are considered homologous body parts. Structures along a body axis, similar but different, are called serially homologous with respect to each other.[/ref] in simpler terms, its body pattern. They are “master” genes because they determine whether a given part will form or not, leaving the details to genes farther down the chain. But such “detail” genes will not function at all without the “master” gene, which therefore regulates quite a large number of genes. Hox genes are sufficiently similar that introduction of mouse Hox genes into a fly can cause the growth of the indicated organ — in fly format. They also control the very different serial structure of snakes.
Homeotic genes occur in clusters. One more amazing fact is that the genes of a cluster are in the same order as that of the body segments they control. It is sufficient to replace the gene in a given cluster, say at the antenna position on a fruit fly, with another, say a leg gene, and a leg develops at the antenna position on the fly. Since the transcription factors coded by such genes can change the cells they regulate into something else, they are called homeotic[ref]Homeosis is the transformation of one organ into another.[/ref] transcription factors and their genes are homeotic genes. The protein domain they express is therefore a homeotic domain, Hox, for short.
Other homeobox families also exist, as we will see a few in a moment. Hox genes are just one family of them.
It is remarkable that quite similar homeodomains have been found in almost all animals. Such conservation of homeobox genes across species shows that embryonic development of most animals, fungi and plants is controlled at some level by approximately the same genes. They must have been around since animals diverged from each other over 500 Mya. The original Hox gene was duplicated and then each copy took on slightly different functions. Subsequent duplications and modifications have led to the diversity of animals today. Comparison of the genes can contribute to building at least a partial tree of life.
Because the homeobox genes code for transcription factors, each of which is used in so many organisms in similar ways, the proteins coded by homeobox genes are constituents of what some biologists call the genetic toolkit. It’s like using a common screwdriver to drive screws in different contexts. Just as one screwdriver serves in many contexts, so does each type of homeobox gene. A homeobox gene is therefore in some way a “master” gene. The toolkit is common to almost all animals, with only little variation from one to another. It contains genes not only for transcription factors, but also for various molecules which are signaling elements. They play important roles in embryonic development, or embryogenesis.
In other animals also, the genes exist in clusters, with the gene order in the cluster corresponding to that of the organism’s parts. Different Hox genes, being similar but slightly different, bind to different regulatory sequences on DNA and therefore regulate different genes. One homeobox protein may regulate many genes and a number of homeobox proteins may work together to refine selection. Because of this possibility of multiple binding, a small change in activation of toolkit genes can bring about a large change in the phenotype. So the genetic toolkit may explain development more simply than if all genes had to be specific to each different part, location and development time of an organism.
Toolkit genes themselves have multiple switches. Switches are the means by which a relatively limited set of toolkit genes may be used differently in different regions, or even different animals, or at different times in embryonic development – which furnishes material for evolution.
The existence of different layers of transcription factors also explains how a small genetic change (in a transcription factor) can bring about a relatively important change in the phenotype of the organism.
A specific bodily environment (liver, heart, blood, …) contains some set of organic molecules specific to that environment. These molecules or a sub-set thereof will serve as transcription factors to activate a particular sub-set of the toolkit. In other words, the environment chooses which tools to use.1 The proteins expressed by toolkit genes will activate or suppress expression of body-part proteins at that place and time.
Environmental molecules ==> toolkit proteins ==> body parts
Each arrow indicates that the object to the left switches on expression of the object to the right.
Some terminology helps to understand the evo-devo literature.
- Transcription factors are proteins and so are not on the DNA string, therefore not on the same molecule as the DNA which is regulated. They therefore are called trans-acting regulatory elements (TRE).[ref]In Latin, “cis” means “this side of” and “trans” means “the other side of”. Think of cis-Alpine (this side of the Alps) and trans-Alpine.[/ref]
- Switches are on the same string of DNA as the regulated gene and are called cis-acting regulatory elements (CRE).
So one can say that TREs bind to CREs to regulate gene expression. Got that?
The following table lists just a few of the homeobox families and the organism components they regulate. As is clear, they regulate quite different structures.
| Protein name | Penotype regulated |
| Hox | body regions (e.g., head, thorax or abdomen) |
| Pax-6 | eyes |
| Distal-less (Dll) | limbs |
| Sonic | organogenesis (tissue patterns) |
| Ulrabithorax (Ubx) | represses insect wing formation |
In all animals, there exist similar gene sequences corresponding to protein domains which are transcription factors for that animal’s version of some phenotype. A Pax-6 gene from a mouse makes an eye form in a fruit fly – a fruit-fly eye, not a mouse eye.
Cell differentiation
Stem cells are those which may split and form any kind of cell.[ref]Usually to make an identical stem cell and another cell, which may or may not be a stem cell.[/ref] But once a specific type of cell is made, it can only do certain things. This is because it no longer has access to the entire recipe book (genome), but only those recipes which it needs. The cell is then said to be differentiated and the process for making it is differential gene expression. Such gene regulation or differentiation depends on the cell’s environment. We have seen an example where the presence of lactose induces the expression of the lac operon.
Gene regulation can fill a book. And cell differentiation can fill another.
Continue with cell division and the cell cycle.